Experiment Description Specification
Terminology
record/entry - collection of fields and attributes with a unique identifier organized together.
Example: “sample_01”: {“type”:”tissue”, “weight”:”30”, “units”:”grams”}
field/attribute/property - piece of data with a unique identifier associated with a record.
Example: weight for a sample entity record.
Overview of Tables
- The data schema developed for MESSES was designed to capture generalized experimental descriptions and data in an abstract way.
The data schema is organized into several tables with a unique record identifier and a flexible collection of fields.
- This schema has the following tables:
entity
protocol
measurement
factor
project
study
Although the tables have been generally described below, there are more details and logic needed to explain the framework.
project
- A project generally refers to a research project with multiple analytical datasets derived from one or more experimental designs.
It is somewhat arbitrary how to categorize a project, but defining one project per group of related analytical datasets derived from a single experiment is a good start.
study
- A study is generally one experimental design or analytical experiment inside of the project.
For instance, if you are testing the effectiveness of a treatment on lung cancer and you try the treatment on both cell lines and mice, the cell line experiment would be one study and the mouse experiment another.
protocol
- A protocol describes an operation or set of operations done on a subject or sample entity.
For example, if mouse organs are frozen in liquid nitrogen and then ground into powder, this could be either 1 protocol to describe both operations or 2 to separate the freezing step and grinding step.
How to break operations up can be an arbitrary decision, but some good rules of thumb are to make operations with significant time between them separate protocols, or if there are measurements associated with an operation.
For example, if the tissue is weighed before and after freezing and before and after grinding, it might be a good idea to have the operations in separate protocols so the weights are clearly associated with each operation.
entity
- Entities are either subjects or samples which are similar to each other and interconnected.
- A subject is something that receives a treatment, or is subjected to different experimental factors.
For example, if you are testing a treatment on a cell line in 6 different petri dishes (1 treatment with 3 replicates and 1 null with 3 replicates), each petri dish would be a separate subject.
- Samples are collected from subjects.
For example, if we revisit the previous example and assume that media is collected from each petri dish, the collected media from each petri dish would be a separate sample.
Samples can also come from other samples. For example, if you take the previously described media samples and take a portion or the entire sample, and then perform several steps before measuring something, such as adding a solvent and/or some other preparation step, the newly modified sample or portion technically becomes a new sample that was derived from the original sample.
- Samples can also become subjects if they are subjected to a treatment.
For example, when a cancer sample is extracted from a person and then pieces are implanted in mice for testing with specific treatments, the mice are subjects with sample parents.
Determining what is a sample and a subject can be confusing, but largely the key is whether or not the thing in question underwent a treatment or has an experimental factor of interest.
Entities must have a “type” field that indicates whether it is a “subject” or “sample”.
measurement
- A measurement is typically the results acquired after putting a sample through an assay or a sophisticated machine such as a mass spec or NMR, as well as any calculations on these results that is used for analysis.
This is what people will typically think of as data while the information in the other tables would be experimental metadata.
factor
- A factor is a controlled independent variable of the experimental design. Experimental factors are conditions set in the experiment. Other factors may be other classifications such as male or female gender.
For example, if there are different treatments done to cell lines this would be a factor.
Each treatment would be a separate protocol, but the different protocols would be 1 factor.
If an experiment also looks at age, race, gender, etc. then these would each be separate factors.
Additional Rules
ID Fields
- Any field for any record that ends in “.id” must be an id to an existing record in that field’s table.
For instance, if the field “protocol.id” : “tissue_extraction” is in a sample record then the protocol table must contain a record with the id “tissue_extraction”.
Similarly, if a record has the “parent_id” field the value must be an existing id in the same table as that record. For example, if “parent_id” : “tissue_extraction” is in a record in the protocol table then the protocol table must contain a record with the id “tissue_extraction”.
Subject/Sample Inheritance Rules
If a sample comes from a sample, it must have a sample_prep type protocol.
If a sample comes from a subject, it must have a collection type protocol.
Subjects should have a treatment type protocol.
Overview of Protocol Types
- There are currently 5 different types of protocols:
treatment
collection
sample_prep
measurement
storage
treatment
- A treatment protocol is used to describe the experimental factors done to subject entities.
For example, if a cell line is given 2 different media solutions to observe the different growth behavior between the 2, then this would be a treatment type protocol.
Each treatment would be a separate protocol that describes the specifics of the solution or other factors.
collection
- A collection protocol is used to describe how samples are collected from subject entities.
For example, if media is taken out of a cell culture at various time points, this would be a collection protocol.
Details such as the time of collection and the weight or volume of collection may be some of the attributes associated with the protocol.
sample_prep
- A sample_prep protocol is used to describe operations done to sample entities.
Typically, these operations would be done or are necessary in preparation for going into a measuring device.
For example, once the cells in a culture are collected, they may be spun in a centrifuge or have solvents added to separate out protein, lipids, etc.
These steps would be a sample_prep protocol.
How to organize such operations into protocols can be arbitrary and is left to the discretion of the creator.
Details such as the concentration of solvents, speed of the centrifuge, or weight of the separated parts may be some attributes associated with the protocol.
measurement
- A measurement protocol is used to describe operations done on samples to measure features about them.
For example, if a sample is put through a mass spectrometer or into an NMR.
Typically, the results of the measurement will be attributes associated with the protocol.
This is also the protocol type used for any analysis or calculations done on the data generated by instruments such as a mass spectrometer.
storage
- A storage protocol is used to describe where things are stored.
This was created mostly to help keep track of where samples were physically stored in freezers or where measurement data files were located on a share drive.
But this protocol should include other storage details like temperature for the physical storage of samples.
Best Practices For Designing Your Experiment Description Data Schema
So far, this document has focused on the mechanics or rules of the experiment description specification, but there are many different ways to create a data schema for the same experiment description. Creating an experimental description data schema is a balance between storing data in a human readable way, storing data in a space saving way (not repeating fields), and storing data in a machine readable way. For the most part, the biggest question to answer is what table or what type of records a piece of information should go into. Some will seem obvious, such as putting a sample’s weight measurement on the sample’s entity record instead of creating a measurement record associated with that sample’s entity record. Others may have several options with advantages and disadvantages. For example, where should you put the name of the raw file generated by a mass spectrometer? On the measurement protocol? On a storage protocol associated with the measured entity? On the entity that was measured? On the measurement records? There can be good reasons for putting the information in different locations and you may have to decide what is best for your situation. This section will try to outline some rules and examples of good ways to construct a data schema based on what the Supported Conversion Formats expect for the conversion command and other factors such as saving space.
You don’t necessarily have to capture absolutely everything about your experiment. Most likely you are doing this in order to upload your data into a public repository, so it is a good idea to understand what information that repository wants or allows and tailor your data schema to that. For instance, if you store some samples in a freezer during the experiment, you may not have to capture the temperature they are stored at because the repository doesn’t have a place for you to give that information to them. However, repository deposition requirements do change. So if it is easy to capture it, it is better to capture it, because you may need it eventually.
Protocols
You may understand every step that was taken in your experimental design, but determining how to group and categorize the steps into protocols can be challenging. It is a good idea to write out everything that was done in an experiment in order to get a measurement. Using a flowchart can be very effective at identifying protocols needed to capture the whole experimental process. This can then be used to help group and categorize what was done into the types of protocols defined by the data schema.
treatment
A treatment protocol will usually be the first protocol and there will often be at least 2 because the control should be considered a separate treatment protocol.
If you did not have controls, but had different subject types then you can create a treatment protocol for each different subject type, even though the procedure is the same. Whether or not this is a good idea is up to you and the data repository you are targeting.
A treatment is usually the procedure that is done to the subjects that induces some kind of change that is of interest to be investigated through measurement. This is different from a sample_prep protocol which describes a procedure done to samples, typically to enable some type of a measurement or to preserve the sample for later use.
Some good information to put in the fields of the protocol are things like dosages or measurable bits of information that differ between treatments. Always make sure to put a good description in the description field. A filename to a document that describes the protocol in much more detail can be given in a filename field.
collection
A collection is only made from a subject, and subjects must receive a treatment. So it is a good idea to start by identifying what the treatment is, which will identify the subjects.
When things are taken from the subjects or if they are sacrificed and transformed, then a collection has happened and a collection protocol is used to indicate that.
The collection protocol should be a protocol for the newly created sample, not the subject it was collected from.
If you collect many different things from a subject, such as many different organs from a sacrificed mouse, you can make a protocol for each different tissue type, or 1 protocol that collects them all.
It is ultimately up to you, but generally it would be separated by procedure. For example, the aforementioned sacrificed mouse would have all of its organs collected at the same time in the same way, so 1 protocol is probably more appropriate. If you collect blood from the mouse at different times as well as when it is sacrificed, then it is probably better to make the blood collection a separate protocol.
Some good information to put in the fields of the protocol are things like the type of sample being collected, such as tissue, plasma, media, etc. Similar collections done at different times can record time points on the protocol itself (requiring a new protocol for each time point), or on the newly acquired sample created from the collection. The latter is what is recommended. Always make sure to put a good description in the description field. A filename to a document that describes the protocol in much more detail can be given in a filename field.
sample_prep
sample_prep protocols are typically procedures done to samples to enable a measurement or preserve a sample for later use.
Most of the steps done to samples will likely fall into this type of protocol.
What can be difficult is deciding which steps to organize under different protocols.
There is generally some middle ground between putting every step under one protocol and making every step its own protocol.
If a set of steps are always done together, then that is a good indication they should be together in one protocol.
If some steps are separated by a significant amount of time or the later steps are not always done, then this is a good indication that the later steps should be part of a separate protocol.
For example, if you harvest mouse organs, freeze them immediately, but only sometimes grind them into powder or the grinding is sometimes done the next day, then making the freezing and grinding separate protocols is a good idea.
Steps done for different purposes are also a good reason to put them in different protocols.
For example, a freezing step may be done to stop metabolic activity, while a step that adds a solvent may be done to separate compounds of interest.
Both of the previous steps are done for the larger purpose of enabling accurate and precise measurements, but have a more specific purpose inside that larger purpose.
Some good information to put in the fields of the protocol would be the amounts or weights of anything added to a sample or taken away if it is the same for each sample. For example, if you add 20uL of a solvent to every sample, then adding that as a field is a good idea. But if you add an amount that is proportional to sample weight or differs between samples then it is probably better to have that as a field on the sample. Other important details or things that can be measured are good targets to add as fields for the protocols. For example, if you add a solvent at a certain temperature, then it may be a good idea to record that temperature. Always make sure to put a good description in the description field. A filename to a document that describes the protocol in much more detail can be given in a filename field.
measurement
measurement protocols are typically the last thing done to a sample in order to quantify something of interest, but if the measurement is non-destructive then there may be more later.
Procedures that operate on entities usually stop after taking a measurement, but additional statistical or analytical steps may continue on the measurement results.
The analysis or statistical steps can be included in the protocol that describes taking the initial raw measurements, or they can be separated into their own measurement type protocols.
There is no rule that says a measurement type protocol cannot be on an entity, but typically only measurement records will have measurement type protocols.
Just because a measurement was taken during a procedure doesn’t mean it has to be a measurement type.
The measurements themselves should be fields on measurement records, but other important information needs to be in the fields of the measurement protocol. Good targets for information to be in the measurement protocol are the software used, the machine used, and specific settings or details about the software or machine, such as version number or the type of chromatography used. Always make sure to put a good description in the description field. A filename to a document that describes the protocol in much more detail can be given in a filename field.
storage
storage protocols are for indicating when something was stored.
This can be an entity or a file.
They were originally created to help keep track of where entities and files were located, but that function is not likely to be useful for depositing into an online repository.
They can still be useful for indicating that an entity was stored somewhere, instead of including it as part of a sample_prep protocol.
Doing this is a good way to highlight specifically that the entity was stored, and how it was stored.
Good information to put on storage protocols are the location of storage, temperature of storage (if relevant), and date of storage. This information can be on the protocol itself, but it might be better for it to be on the entity the protocol is associated with. Determine what is best for your situation.
Entities
Entities and protocols have an interdependent relationship. Entities must have at least 1 protocol associated with them, and protocols describe what was done to the entities. Two of the biggest challenges are determining how to break up the experiment procedures into protocols and how to create entities from experiment subjects and samples. Due to their interdependence, this can be confusing. The following are some tips that can help:
If any sort of collection takes place (blood, tissue, media, etc.), then the entity it came from must be a subject and the collected entity is a sample that must have a collection protocol associated with it.
The subject must be the parent_id of the sample.
If a sample is aliquoted into different portions, then each portion becomes a new separate entity and those entities parent_id’s are the original sample.
Portions that do not go on to be measured may be omitted in a data deposition. Use your own judgment.
New entities should not be created for every individual sample preparation step, but major steps/procedures that enable a measurement.
New derivative samples should be used to describe the procedures needed to craft a sample for a specific analytical measurement.
If the samples are only used for a single type of analytical measurement, then the protocol needed to create the actual sample used for the measurement could just be part of the list of protocols for the sample.
But if the samples need to be split and processed into separate samples for different types of analytical measurements, then this will need derivatives samples that have the associated specific sample_prep protocol.
If there is a significant amount of time between sample_prep protocols, or the sample is stored for a period of time, then it may be a good idea to create a new derivative entity after it is taken out of storage or after the delay between steps.
If there is a protocol that is not always done, or is not done to all of the samples, then that is a good indication to create a new derivative entity when that protocol is done on an entity.
It can be useful to think about what real world physical samples existed during the experiment. For example, if you treat a mouse and tissue is collected from it that then has other procedures applied to make it measurable, then it can be helpful to create a diagram depicting the mouse, each tissue, and each test tube after that. This diagram can then be filled in with protocols connecting one to the next. An example is shown below.
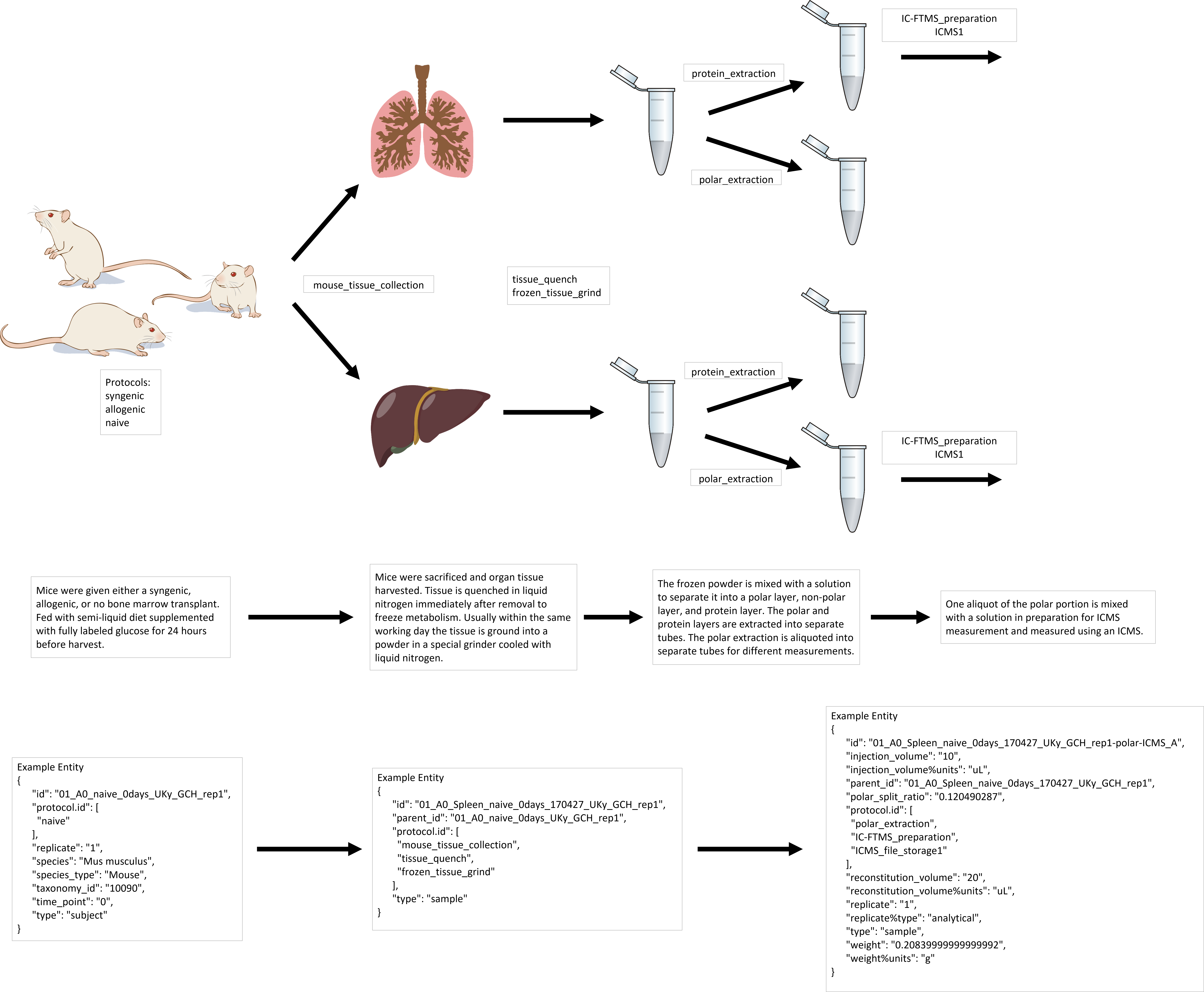
Measurements
There are usually many measurements taken during an experiment, but not all of them need to go in the “measurement” table. Generally, experiments result in one or more end point measurements and these are what will go in the “measurement” table. Assuming you are uploading to a repository that repository might also influence what goes in the “measurement” table. For example, a mouse experiment might use a mass spectrometer and a nuclear magnetic resonance spectrometer to take measurements of metabolites in mouse tissue. The purpose of the experiment was to be able to analyze and compare these metabolite measurements, so they are the end point measurements. You also want to upload them to the Metabolomics Workbench, which expects metabolite measurements from MS and NMR arranged in a specific way. These are both good reasons to have them be in the “measurement” table.
Analysis and transformations of the measurements should also go in the same record as the measurement. For example, if you identified a peak in a mass spectrometry spectra as glucose and put the peak area measurement in the “measurement” table, then you would also want to put any normalization calculations with it in the same measurement record. So the glucose record would have a field for raw_intensity and corrected_raw_intensity.
You have decided what should be in the “measurement” table, but there is still the question of how exactly to create the records. Sticking with our mouse experiment example, you could create 1 record per metabolite and have the measurements for each sample as a separate field (ex sample1_raw_intensity, sample2_raw_intensity, etc.). What the convert command expects, however, is a separate record for each metabolite-sample combination. Ex:
"measurement": {
"Glucose-Sample1": {
"assignment": "Glucose",
"assignment%method": "database",
"compound": "Glucose",
"concentration": "0",
"concentration%type": "calculated from standard",
"concentration%units": "uM",
"corrected_raw_intensity": "8447352.89211",
"corrected_raw_intensity%type": "natural abundance corrected peak area",
"entity.id": "Sample1",
"formula": "C6H12O6",
"id": "Glucose-Sample1",
"intensity": "13664945.509939667",
"intensity%type": "natural abundance corrected and protein normalized peak area",
"intensity%units": "area/g",
"normalized_concentration": "0",
"normalized_concentration%type": "protein normalized",
"normalized_concentration%units": "uMol/g",
"protocol.id": "ICMS1",
"raw_intensity": "7989221.83386388",
"raw_intensity%type": "spectrometer peak area"
},
"Glucose-Sample2": {
"assignment": "Glucose",
"assignment%method": "database",
"compound": "Glucose",
"concentration": "0",
"concentration%type": "calculated from standard",
"concentration%units": "uM",
"corrected_raw_intensity": "2885161.33083",
"corrected_raw_intensity%type": "natural abundance corrected peak area",
"entity.id": "Sample2",
"formula": "C6H12O6",
"id": "Glucose-Sample2",
"intensity": "6235697.006728272",
"intensity%type": "natural abundance corrected and protein normalized peak area",
"intensity%units": "area/g",
"isotopologue": "13C0",
"isotopologue%type": "13C",
"normalized_concentration": "0",
"normalized_concentration%type": "protein normalized",
"normalized_concentration%units": "uMol/g",
"protocol.id": "ICMS1",
"raw_intensity": "2728688.40604858",
"raw_intensity%type": "spectrometer peak area"
},
...
}
We have found this form to be the best practice. Note that not every field shown is required for the convert command. To know what is required by default, or to modify it to fit your situation, it is recommended to read the Conversion Directives and Supported Conversion Formats sections, and use the “save-directives” sub-command for the convert command to investigate the specific fields required for your repository upload. Also notice the “field%type” and “field%units” attribute fields for the measurement values. Attributes are the recommended way to indicate additional information about a measurement value.
Useful Patterns
The following are some problems and ways to deal with them.
Connecting Records to File Names
One common problem is the need to associate raw files with both the sample that was measured to generate them and the measurements pulled from them. There are a few ways to do this and they all have advantages and disadvantages. You should choose a way that is best for your situation.
Add a data_file field directly to the measurement record along with the recommended entity.id field.
This makes it extremely easy to see which file the measurement came from, but that field is repeated a lot, and it is cumbersome to figure out which file goes with which entity.
Create a storage file protocol for each raw file. The protocol would have fields for the file name or path, an entity.id field that points to the entity it is associated with, and a protocol.id field that would point to the appropriate measurement protocol.
This protocol could be a protocol for the measurement record or the entity that was measured.
This repeats the raw file field less, but requires the creation of many protocols that will likely dominate the protocol table, all to hold a raw file name or path.
It is also not as straightforward to determine which raw file goes with which measurement, since you have to match up the entity.id and/or protocol.id from the measurement record with those on the storage protocol.
A similar approach to the previous option is to create separate measurement protocols for each raw file and put the file name or path as a field in the protocol. This has similar downsides to create storage protocols, but the measurement protocols have more significant fields that will be repeated many times.
This is easier to determine which raw file goes with which measurement, but takes a little more work to link raw files with samples unless you also include an entity.id field on the protocol.
Add a data_files list field to both the entity and measurement protocol. Assuming you have an entity.id field on your measurement records then you can cross reference the lists and what they have in common is the file for the measurement.
This has a lot less repetition, but does require unique raw file names (this can be enforced using the protocol-dependent schema).
Although determining which file goes with which measurement can be done by cross referencing the lists, it is more complicated than simply looking up a field.
Add a data_files list field and a data_files%entity_id onto the measurement protocol. The lists would be linked such that the positions in each list correspond to one another. For example, if you have a data_files list on a measurement protocol, [file1, file2], and a data_files%entity_id list, [entity1, entity2], then file1 would be associated with entity1 and file2 with entity2 because they are in the same numerical position in their respective lists.
This does require the lists to be the same length and leaves open the possibility of the lists getting out of sync, but if you use the automation tags with the extract command you can reduce the likelihood of that happening.
Determining which file goes with which measurement just requires matching the entity.id field of the measurement record with a value in the data_files%entity_id list.
Determining the file that goes with the measurement or sample is more complicated than simply looking up a field, but there is much less repetition of redundant information.
Compared with option 4 there is also the advantage that both lists are in 1 table and record.
Add a data_files list field and a data_files%measurement_protocol onto the entities. The lists would be linked such that the positions in each list correspond to one another. For example, if you have a data_files list on an entity, [file1, file2], and a data_files%measurement_protocol list, [protocol1, protocol2], then file1 would be associated with protocol1 and file2 with protocol2 because they are in the same numerical position in their respective lists.
This does require the lists to be the same length and leaves open the possibility of the lists getting out of sync, but if you use the automation tags with the extract command you can reduce the likelihood of that happening.
Determining which file goes with which measurement just requires matching the protocol.id field of the measurement record with a value in the data_files%measurement_protocol list.
Determining the file that goes with the measurement is more complicated than simply looking up a field, but there is much less repetition of redundant information.
Compared with option 4 there is also the advantage that both lists are in 1 table and record.
This was a decision we had to make for the data schema that would be expected when converting to the mwtab format. Ultimately, we went with option 5. To see how this works you can look at the mwtab examples in the examples folder of the GitHub repository. Look at the #automate sheets of the measurements Excel files to see how you can easily add these fields in a way that will reduce the chance of having misaligned lists. Also check out the protocol-dependent_schema files to see how to enforce unique file names.